(From left) Mr Duan Xin, Dr Lin Xudong, Dr Shi Peng, Professor Cheng Shuk Han and Dr Wang Xin.
A research team led by City University of Hong Kong (CityU) has found a shortcut for developing new drugs which can potentially reduce time and costs by sorting out high potential candidates out of a long list of chemical compounds, with an accuracy of around 50%.
This breakthrough in neuropharmacology came following five years of collaborated research by CityU’s Department of Biomedical Engineering (BME) and Department of Biomedical Sciences (BMS), and Harvard Medical School. The research is published in the scientific journal Nature Communications and titled “High-throughput Brain Activity Mapping and Machine Learning as a Foundation for Systems Neuropharmacology”.
The research, led by Dr Shi Peng, Associate Professor of BME, provides a platform to predict compounds that have the potential to be developed into new drugs to treat brain diseases. It can help speed up the new drug discovery process and save costs.
“Even a 1% increase in the drug development success rate would make a huge difference for central nervous system (CNS) disorder patients,” Dr Shi explained.
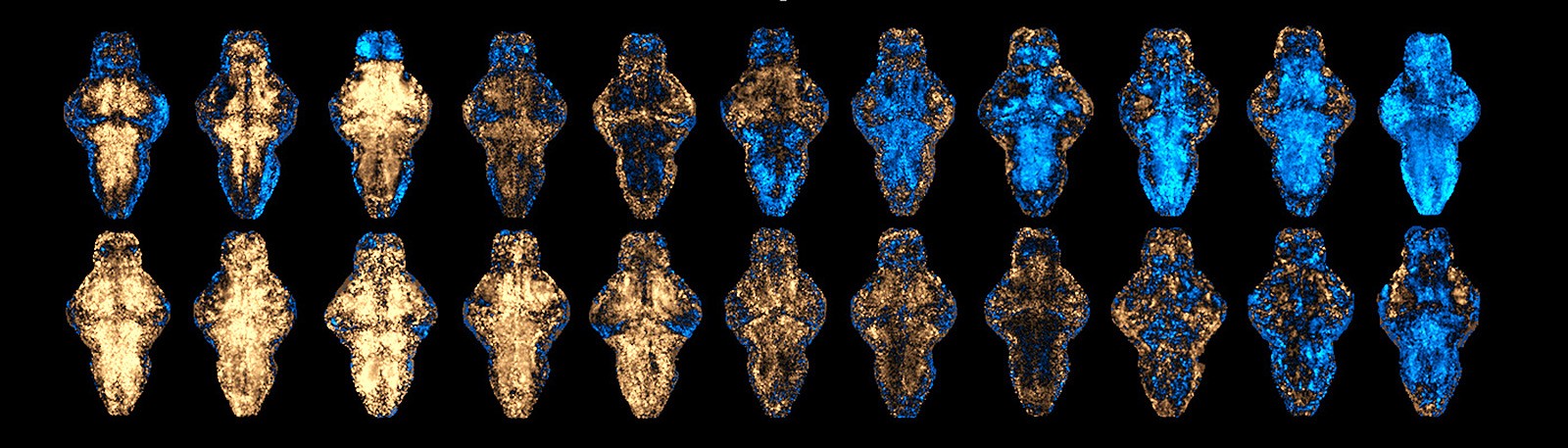
The study used zebrafish, a small vertebrate animal to conduct whole-brain activity mapping, which shows how the brain or the CNS react to the drugs. The setup was streamlined with innovative system to enable large-scale experiments.
“We used robotics, microfluidics and hydrodynamic force to trap and orient an awake zebrafish automatically in 20 seconds, which took 20 minutes in the past. In this way, we can carry out imaging for many zebrafishes in one go. More importantly, our platform can immobilise the fish without anaesthesia, thus avoiding interference,” Dr Shi explained.
The team first built a reference library of brain activity maps for 179 existing CNS drugs. They generated the maps from the brains of thousands of zebrafish larvae, each treated with a clinically used CNS drug. The maps showed the corresponding brain regions that reacted to those drugs. The team then classified these drugs into 10 physiological clusters based on the intrinsic coherence among the maps by machine learning algorithms.
With the reference library in hand and in close collaboration with Dr Wang Xin, Assistant Professor of BMS at CityU, and Dr Stephen Haggarty, Associate Professor at Harvard Medical School, the team went on to carry out information analysis and employed machine learning strategy to predict the therapeutic potential of 121 new compounds.
The machine learning strategy predicted that 30 out of those 121 new compounds had anti-seizure properties. To validate the prediction, the research team randomly chose 14 from the 30 potential anti-seizure compounds to perform behavioural tests with induced seizure zebrafishes.
The result showed that 7 out of 14 compounds were able to reduce the seizures of the zebrafish without causing any sedative effects, implying a prediction accuracy of around 50%. “With this high-speed in vivo drug screening system combined with machine learning, we can provide a shortcut to help identify compounds with significantly higher therapeutic potentials for further development, hence speed up the drug development and reduce the failure rate in the process,” Dr Shi said.
The first co-authors of the paper are Dr Lin Xudong, Research Associate of BME, and Mr Duan Xin, Research Associate of BMS. Other authors include Professor Cheng Shuk Han of BMS, Mr Chan Chung-yuen, BME graduate, and Ms Chen Siya, former Research Associate of BME.